Protein |
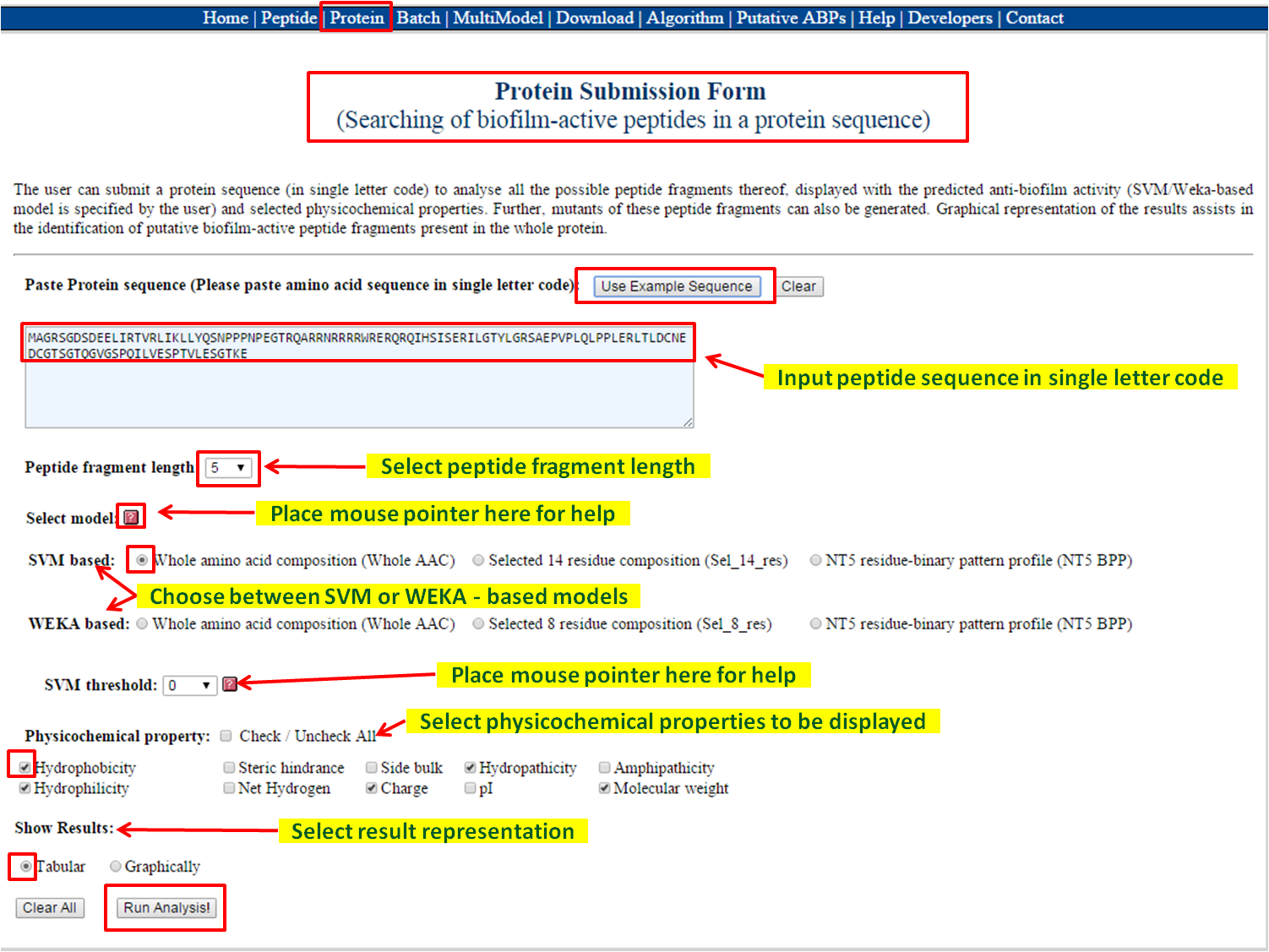
| There user can identify anti-biofilm regions from the protein of interest. Possible peptides are generated from the protein using overlapping window concept. The snapshot given below gives a very brief idea about this module. | |
 |
The results from Protein module may be generated in following two modes:
|
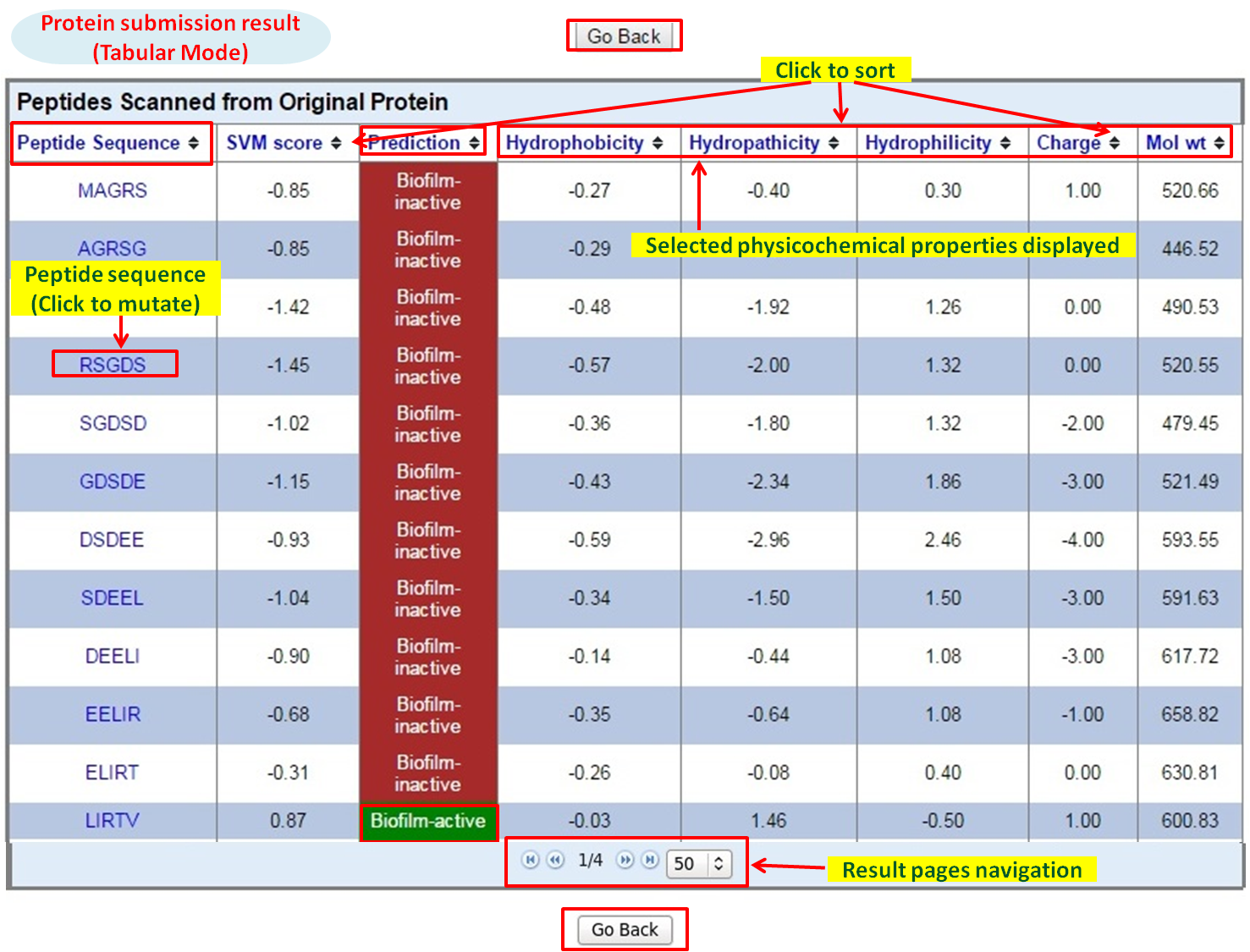
Tabular Mode |
The brief description is given below:
- Paste the protein (in single letter code, single line sequence only) in the input box. After submission, all possible peptides are generated containing putative anti-biofilm ability (in the form of SVM score / Weka probability) with physicochemical properties.
- Results are displayed in tabular form (having desired physicochemical properties choosen by the user).
- Mutants of the fragments with anti-biofilm properties can be generated further by just clicking on the fragment. The later will help to generate best putative anti-biofilm peptides.
|
 |
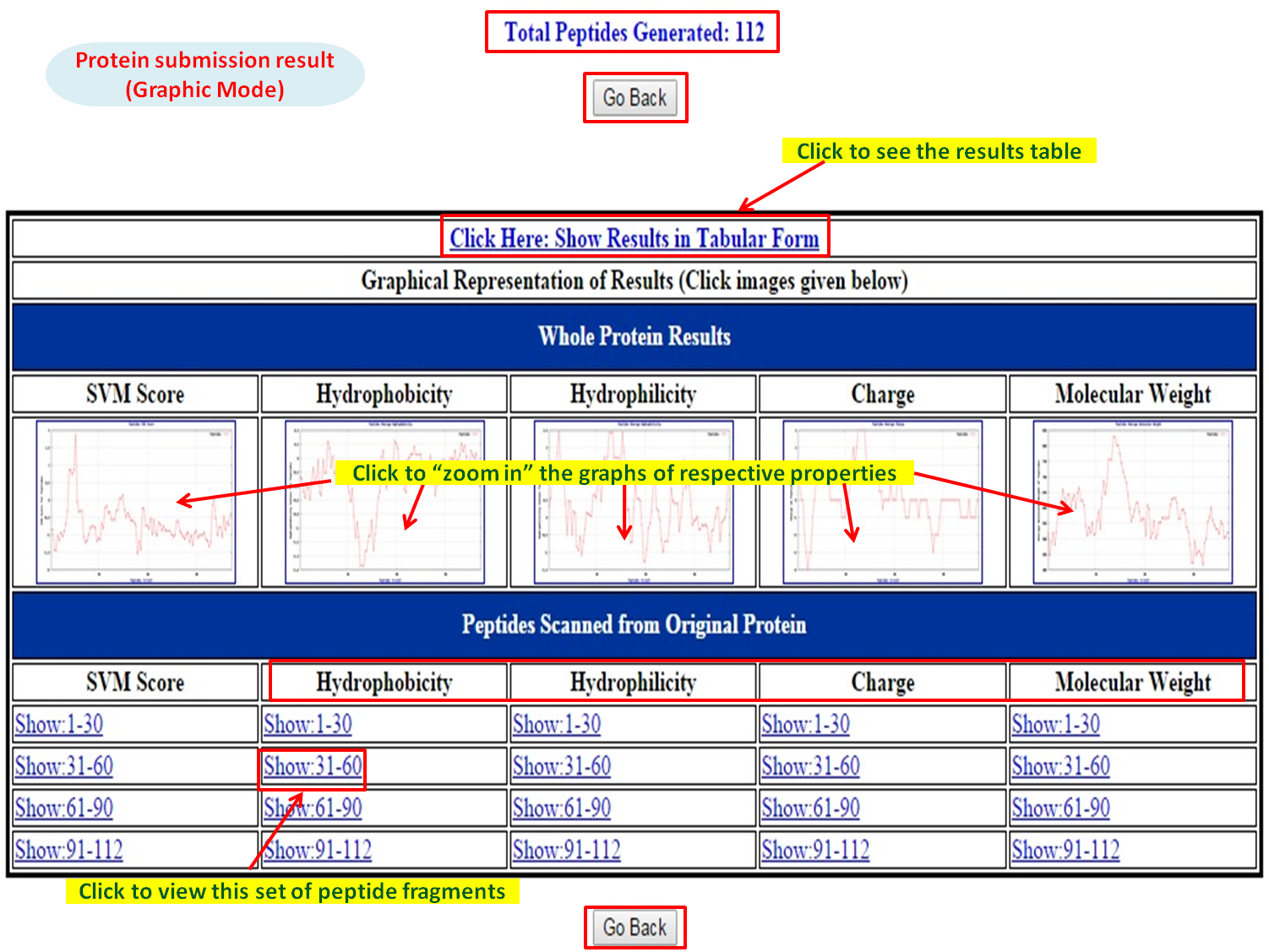
| Graphic Mode |
| The results are displayed in two patterns, one showing the whole protein (in terms of all overlapping peptide fragments) prediction results (SVM score / Weka probability) accompanied with four physicochemical properties (i.e., Hydrophobicity, Hydrophilicity, Charge, Molecular weight). Clicking upon a particular physicochemical property e.g., on the basis of SVM score (at a particular threshold), no. of peaks will decide possible anti-biofilm peptides present in protein sequence. | |
| The other pattern is displayed for fragments which are generated from the submitted protein. The predicted bioactivity value (SVM score / Weka probability) accompanied with physicochemical properties (Hydrophobicity, Hydrophilicity, Charge, Molecular weight) and the peptide fragments for that protein are displayed. On clicking the peptide fragments, the predicted bioactivity scores / properties are displayed in graphical form. |
 |





